
|
||
| Joe Jaklevic (far right) and his colleagues in the Engineering Division's Bioinstrumentation Department. | ||
Today, synchrotron light from facilities such as Berkeley Lab's Advanced Light Source may make it possible to use protein crystals as small as 50 microns (50 millionths of a meter) in length.
The crystals themselves may also become easier to grow, thanks to a unique robotic system designed and built by Joseph Jaklevic, head of Engineering Sciences, and his colleagues in the Engineering Division's Bioinstrumentation Department.
"The idea for a high-throughput combinatorial approach to crystal growth came from Peter Schultz," says Jaklevic. "The basic idea is that, instead of having to plod through all the hundreds of ways you might get a protein to crystallize, you more or less try 'em all at once."
Schultz pioneered combinatorial methods as a member of the Lab's Materials Sciences Division; he recently became head of the Novartis Institute for Functional Genomics in La Jolla, California. He and his colleague Raymond Stevens of the Lab's Physical Biosciences Division saw the combinatorial approach as a natural solution to the challenge of growing protein crystals.
That's because "biologists really have no idea what the best conditions are for growing crystals of a new protein," says Derek Yegian, a member of the team that built the new robotic system. "Different proteins precipitate out of solution and grow at different rates-or don't grow at all-depending on the solution's acidity, temperature, concentrations of salts, and lots of other variables. "
 |
|
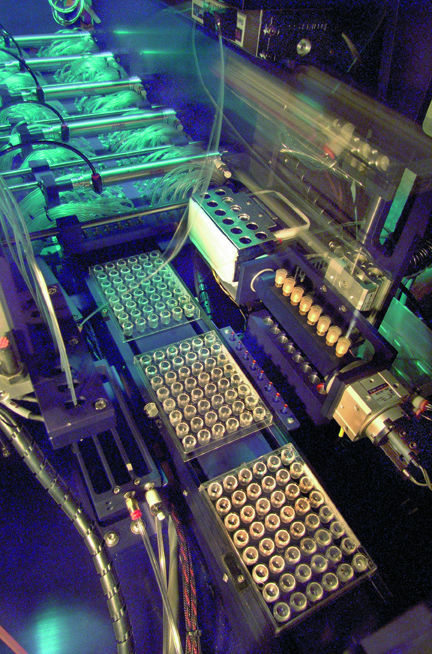
| The innovative robot above, designed and built by Joe Jaklevic and his colleagues in the Engineering Division's Bioinstrumentation Department, can automatically grow crystals of a novel protein by screening 480 different growth solutions at once. | |
Only the very purest proteins will crystallize, and pure protein is expensive; even common commercial proteins can cost hundreds of dollars a gram. Often hundreds of combinations of variables must be tried before a novel protein can be crystallized from solution.
Most trial solutions are prepared by hand at the rate of about 30 an hour, typically requiring one to 10 microliters of pure protein for 50 to 100 "coarse-screening" trials; whether a particular solution yields a crystal is apparent only days or weeks later.
"Manual methods are slow and error-prone," says Yegian, and although some steps have been automated within the past few years, "commercial robots are not much better." With the Bioinstrumentation Department's new robotic system, however, once a target protein has been chosen, 480 different variations of growth solution can be coarse screened all at once, each in its own tiny reservoir.
A set of trials starts with 10 empty, transparent plastic cassettes. As each cassette is loaded from a stacker, the robot lifts its lid, and 48 needles simultaneously coat the lips of the 48 tiny bowl-shaped wells inside with a thread of grease.
Next the wells are half-filled with growth solution, called the "mother liquid," by syringes fed by banks of cylinders; each bank of 10 holds 48 different solutions, varying by types of salts, buffers, and so on.
Meanwhile, at a separate station, a syringe deposits a mixture of the protein and the appropriate mother liquid on each of 48 circular transparent cover slips. As the cassette is stepped through, each row of eight slips is inverted and sealed over the corresponding wells. The robot needs six minutes to set up and seal the 48 cover slips for a cassette, which sets the rate of the entire coarse-screening preparation process.
 |
|
The cassettes are automatically stored at constant temperature after filling. Water slowly diffuses from the hanging drop into the mother liquid in the reservoir, and the concentration of protein increases. Crystals precipitate in some of the reservoirs.
"The smaller the drop size, the quicker the crystallization," Jaklevic explains. "We're trying to get crystallization with a few nanoliters of protein instead of microliters-a reduction by a factor of a thousand. We will see crystals within a few hours to a few days."
A high-resolution ccd camera checks the cassettes twice a day and detects the growth of even tiny crystals in the transparent reservoirs; reservoirs showing crystals can be returned to the sample preparation unit to begin a second cycle, under conditions finely tuned for optimum growth. Solutions that don't produce good results are rejected. The preliminary system has been successfully tested for reliability, consistency, and speed on commercially available proteins and on novel proteins from the laboratories of Schultz and Stevens. It includes hardware built from scratch or adapted by the Bioinstrumentation Department and uses original software to control the numerous delicate steps in the operation; Jaklevic and his colleagues are working to replace or improve some off-the-shelf components. Before long, they hope to see the full system in regular use at the Advanced Light Source and elsewhere.
 |
|
| Most protein crystals must be larger than a tenth of a millimeter to be useful in crystallography. The ALS will be able to use crystals as small as 50 microns. | |
Jaklevic and his department have designed, built, and modified numerous ingenious robots used in the life sciences, including devices for the Lab's Human Genome Center credited with proving several years ago that prodigious rates of automated gene sequencing were possible with no loss of accuracy. Recently the Bioinstrumentation Department built the second-generation microdot arrayers used by Life Sciences Division researchers to study gene expression in heart disease. "To accommodate the enormous growth in the biological sciences at Berkeley Lab we need to exploit the Lab's traditional multidisciplinary strengths," says Jaklevic. "We hope biologists will see how much they can benefit by making use of engineers. We have a lot of engineers here who can 'speak biology.'" - end -
| < Research Review | Top ^ | Next > |