 |
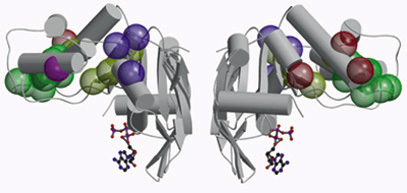
| ABC transporter molecules such as these are responsible for carrying substances back and forth across the inner membranes of cells. Members of the ABC family include a cystic fibrosis regulator and a multidrug resistance protein. |
|
 |
Cystic fibrosis is the most common fatal genetic disease in the United States today, occurring in approximately one out of every 3,300 live births. Now, scientists have obtained their first three-dimensional look at a member of a large family of proteins that play a central role in the development of cystic fibrosis and are also known to block the therapeutic effects of medications. The structure of the protein was solved by a team of researchers at Berkeley Lab and the University of California at Berkeley. Sung-Hou Kim, a chemist with Berkeley Lab's Physical Biosciences Division and a professor with UCB's Chemistry Department, and Giovanna Ferro-Luzzi Ames, a professor in UCB's Molecular and Cell Biology Department, led the research team, which used intense beams of x-rays produced at Berkeley Lab's Advanced Light Source (ALS) to solve the structure of a protein called HisP.
HisP is a "conserved subunit" of a family of proteins known as ATP-binding cassette (ABC) transporters. ABC transporters are responsible for carrying substances back and forth across the inner membranes of cells. Among the many medically significant proteins in the ABC transporter family are the cystic fibrosis transmembrane regulator (CFTR) and a multidrug resistance protein (MDR) called P-glycoprotein.
Cystic fibrosis is caused by mutations in the CFTR gene that result in defective CFTR proteins. MDR proteins are the bane of the medical community because they counteract the effects of pharmaceutical drugs, forcing doctors to increase prescribed dosages in order to obtain desired results.
"Cystic fibrosis occurs when the ABC transporters are not working well enough, and multidrug resistance occurs when ABC transporters are working too well," says Kim. "With our 3-D crystal structure, we have provided a structural basis for understanding the properties of ABC transporters."
The HisP protein that Kim and his colleagues imaged comes from the recently completed Escherichia coli genome. The scientists resolved the protein's crystal structure to 1.5 angstroms, a level of detail made possible by the quality of the x-ray beams and instrumentation available at the ALS' Macromolecular Crystallography Facility. Once they had a 3-D image of their HisP protein, the research team was able to correlate structural details with the biochemical, genetic, and biophysical properties of wild-type and mutant HisP proteins, as well as with some of the known mutants of CFTR and MDR proteins.
"Our findings indicate that HisP is a good model for the ATP-binding domain of ABC transporters in general," says Kim. "It could be used to better understand and perhaps treat cystic fibrosis or to design ways to inhibit multidrug resistance."
Co-authoring the Nature paper with Kim and Ames were Li-Wei Hung, Iris Xiaoyan Wang, Kishiko Nikaido, and Pei-Qi Liu.
Siberian Journey
 |
| The research vessel Persei carried Tamas Torok and his Siberian colleagues across Lake Baikal in search of unknown organisms |
|
 |
If "Vector" sounds like a code name in a James Bond movie, that's not a bad guess, considering that Vector was a secret laboratory and production facility in Siberia, as big as a small city but not on any map, which specialized in research on biological warfare. These days, however, Vector and Berkeley Lab scientists are working together under the Initiatives for Proliferation Prevention (IPP), a Department of Energy program established after the collapse of the Soviet Union to help keep former Soviet defense researchers gainfully-and peacefully-employed.
Late last year Tamas Torok of the Life Sciences Division returned from a visit to Vector and the Lake Baikal region, where in collaboration with Vector microbiologist and institute director V.E. Repin he undertook for ancient and as yet unknown microorganisms with novel medical and biotechnological potential.
Torok's Siberian journey began at Irkutsk, then went on to Ulan Ude and beyond. He traveled by vintage plane-which makes the circuit from Samara to Novosibirsk to Irkutsk just twice a week-then by train, and finally by off-road van, 10 hours over nonexistent roads through the snow and mud of mountains and forests, to the "Saint's Nose" peninsula on Lake Baikal's southeastern shore, dragging his entire laboratory with him in two ice chests.
"The only time I lost my temper was when the conductor wouldn't let me on the train with my chests," Torok recalls. "My Russian colleagues couldn't persuade her, but English somehow did the trick." The ice chests nearly filled his upper bunk, and Torok was reconciled to spending the 10-hour overnight ride clinging to a strip of bedding a few inches wide, until a Buryat man insisted on moving one of the chests into his own bunk.
Torok went to Lake Baikal because it is an isolated environment with extraordinarily diverse aquatic life-some 1,500 species, 85 percent of which have turned out to be unique (including a fresh-water seal!). The likelihood that one could find unusual microorganisms was therefore high. Baikal is not only the biggest lake in the world, with 20 percent of the Earth's fresh surface water-more water than all the Great Lakes combined; it is the world's oldest as well, over 30 million years old. The lake floor averages a mile down, but that floor consists of sediments which fill a rift in the Earth's crust more than five miles deep.
From rented research vessels Torok took samples of deep lake water, seeking unknown organisms adapted to the cold. From hot springs in the surrounding region he took samples which may contain unknown organisms with heat adaptations. Enzymes responsible for resistance to temperature and other extremes, such as acidity, have great potential in manufacturing processes.
A third source of unique specimens came as an unexpected bonus.
"For several years the International Baikal Drilling Project, a consortium of U.S., Japanese, European Community, and Russian scientists, has been taking core samples from sediments in different parts of the lake," Torok explains. "Layers of sediment can be dated almost like tree rings, and by looking at remains of aquatic life and other deposits, they get an indication of how the climate has changed over millions of years.
"The core samples are divided up between the local and international scientists-all kinds of scientists, except microbiologists," says Torok. "When we met, both parties saw an incredible opportunity." Torok was given samples from cores drilled in January of 1998 to bring back to the Lab for analysis. In February of this year he went back for more core samples. Now CEB's lab is chock full of microorganic treasures awaiting analysis.
A scientific success, the trip also left Torok deeply impressed with the Siberians. "Most of the people are so poor they have no reason for dishonesty," he says. "If someone catches a fish, they divide it up right there. In a cabin without running water or beds, they made me sleep on the table; it was a place of honor. They share everything they have. They are so human. And so helpful."
Joint Genome Institute Dedicates New Production Sequencing Facility
 |
|
|
 |
It is the biological equivalent of the race to the moon. A monumental effort joined by researchers across the globe to sequence the three billion units of the human genome, the genetic code that is crucial to understanding human development and countless human diseases with genetic roots. The goal is to achieve a "working draft" of the human genome by next March, and a completed genome by 2003.
With this daunting task in mind, a host of people, incuding Energy Secretary Bill Richardson and more than 250 researchers, local officials and national laboratory representatives, met in Walnut Creek, California, to dedicate the Joint Genome Institute's (JGI's) newest addition,
the Production Sequencing Facility. The new facility will ultimately be occupied by 200 researchers working in three shifts around the clock.
Founded in 1996, JGI is a consortium of scientists, engineers and support staff from the Department of Energy's Lawrence Berkeley, Lawrence Livermore and Los Alamos national labs. Along with a handful of other sequencing facilities in the United States, it is in a race to determine the three billion paired "letters" that comprise the human genome. This global effort-the largest biological undertaking in history-promises untold opportunities to understand the basic molecular underpinnings of life and to improve human health.
"You are the cartographers of the modern day, mapping uncharted terrain, and making easier the life journey for generations to follow," Secretary Richardson said, commending the workers who have dedicated themselves to the historic project.
"We are now finally ready to finish the task at warp speed," Elbert Branscomb, director of the JGI, added. "We will soon be turning out several hundred million letters of genetic text a year."
Martha Krebs, director of DOE's Office of Science and architect of the three-lab collaboration that became the JGI three years ago, emphasized the value of partnerships to the success of the program and cited four individuals for their "inspired leadership" in helping to make the JGI a reality. Among them was Berkeley Lab Deputy Director of Research Pier Oddone.
All the sequence information generated by the JGI is released to public view daily, so the scientific community has free and immediate access to this burgeoning data resource. The benefits of the genome project are already coming to light. Of the 23 pairs of human chromosomes, the JGI's sequencing enterprise targets chromosomes 5, 16 and 19-regions of the human genetic library that have already revealed many new genes, including several already known to be involved in human diseases such as diabetes, cardiovascular disease, asthma, schizophrenia, and various forms of cancer.
|



![]()